Practical - Machine Learning Project#
In this we are going to apply different machine learning models on medical cancer dataset
Import Libraries#
# import Libararies
import pandas as pd
import numpy as np
from numpy import unique
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
import missingno as msno
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
#----- Import Tensorflow modules -----------#
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Dropout
from tensorflow.keras.callbacks import EarlyStopping
# Set plotly as backend for plotting through seaborn in sns l option
pd.options.plotting.backend = "plotly"
Data upload#
# Data upload
meso_data = pd.read_csv("/content/drive/MyDrive/Mesothelioma/Mesothelioma_data.csv")
meso_data.head()
| age | gender | duration of asbestos exposure | duration of symptoms | dyspnoea | ache on chest | weakness | habit of cigarette | cell count (WBC) | hemoglobin (HGB) | ... | pleural lactic dehydrogenise | pleural protein | pleural albumin | pleural glucose | dead or not | pleural effusion | pleural thickness on tomography | pleural level of acidity (pH) | C-reactive protein (CRP) | diagnosis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 47 | 1 | 20 | 24.0 | 1 | 1 | 0 | 2 | 9 | 1 | ... | 289 | 0.0 | 0.00 | 79 | 1 | 0 | 0 | 0 | 34 | 1 |
| 1 | 55 | 1 | 45 | 1.0 | 1 | 1 | 1 | 3 | 7 | 0 | ... | 7541 | 1.6 | 0.80 | 6 | 1 | 1 | 1 | 1 | 42 | 1 |
| 2 | 29 | 1 | 23 | 1.0 | 0 | 0 | 0 | 2 | 12 | 1 | ... | 480 | 0.0 | 0.00 | 90 | 1 | 0 | 0 | 0 | 43 | 2 |
| 3 | 39 | 1 | 10 | 3.0 | 0 | 1 | 1 | 0 | 14 | 1 | ... | 459 | 5.0 | 2.80 | 45 | 1 | 1 | 0 | 0 | 21 | 1 |
| 4 | 47 | 1 | 10 | 1.5 | 1 | 1 | 0 | 3 | 4 | 0 | ... | 213 | 3.6 | 1.95 | 53 | 1 | 1 | 0 | 0 | 11 | 1 |
5 rows × 27 columns
EDA#
meso_data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 324 entries, 0 to 323
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 324 non-null int64
1 gender 324 non-null int64
2 duration of asbestos exposure 324 non-null int64
3 duration of symptoms 324 non-null float64
4 dyspnoea 324 non-null int64
5 ache on chest 324 non-null int64
6 weakness 324 non-null int64
7 habit of cigarette 324 non-null int64
8 cell count (WBC) 324 non-null int64
9 hemoglobin (HGB) 324 non-null int64
10 platelet count (PLT) 324 non-null int64
11 sedimentation 324 non-null int64
12 blood lactic dehydrogenise (LDH) 324 non-null int64
13 alkaline phosphatise (ALP) 324 non-null int64
14 total protein 324 non-null float64
15 albumin 324 non-null float64
16 glucose 324 non-null int64
17 pleural lactic dehydrogenise 324 non-null int64
18 pleural protein 324 non-null float64
19 pleural albumin 324 non-null float64
20 pleural glucose 324 non-null int64
21 dead or not 324 non-null int64
22 pleural effusion 324 non-null int64
23 pleural thickness on tomography 324 non-null int64
24 pleural level of acidity (pH) 324 non-null int64
25 C-reactive protein (CRP) 324 non-null int64
26 diagnosis 324 non-null int64
dtypes: float64(5), int64(22)
memory usage: 68.5 KB
meso_data.isnull().sum().plot(kind="bar")
meso_data.describe().plot()
print(meso_data.diagnosis.value_counts())
print("======================================================")
px.pie(meso_data, hole= 0.6, names="diagnosis", color= "diagnosis")
1 228
2 96
Name: diagnosis, dtype: int64
======================================================
# wide to long data frame
HD_df_long = meso_data.melt( id_vars="diagnosis")
HD_df_long.head()
| diagnosis | variable | value | |
|---|---|---|---|
| 0 | 1 | age | 47.0 |
| 1 | 1 | age | 55.0 |
| 2 | 2 | age | 29.0 |
| 3 | 1 | age | 39.0 |
| 4 | 1 | age | 47.0 |
fig = px.box(HD_df_long, y= "value", color = "diagnosis", facet_col= "variable", notched= True)
fig.update_xaxes(tickangle=45)
fig.show()
meso_data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 324 entries, 0 to 323
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 324 non-null int64
1 gender 324 non-null int64
2 duration of asbestos exposure 324 non-null int64
3 duration of symptoms 324 non-null float64
4 dyspnoea 324 non-null int64
5 ache on chest 324 non-null int64
6 weakness 324 non-null int64
7 habit of cigarette 324 non-null int64
8 cell count (WBC) 324 non-null int64
9 hemoglobin (HGB) 324 non-null int64
10 platelet count (PLT) 324 non-null int64
11 sedimentation 324 non-null int64
12 blood lactic dehydrogenise (LDH) 324 non-null int64
13 alkaline phosphatise (ALP) 324 non-null int64
14 total protein 324 non-null float64
15 albumin 324 non-null float64
16 glucose 324 non-null int64
17 pleural lactic dehydrogenise 324 non-null int64
18 pleural protein 324 non-null float64
19 pleural albumin 324 non-null float64
20 pleural glucose 324 non-null int64
21 dead or not 324 non-null int64
22 pleural effusion 324 non-null int64
23 pleural thickness on tomography 324 non-null int64
24 pleural level of acidity (pH) 324 non-null int64
25 C-reactive protein (CRP) 324 non-null int64
26 diagnosis 324 non-null int64
dtypes: float64(5), int64(22)
memory usage: 68.5 KB
px.scatter_matrix(meso_data,
dimensions= ["age", "duration of asbestos exposure", "duration of symptoms", "dyspnoea", "ache on chest", "weakness", "habit of cigarette", "cell count (WBC)" ],
color = "diagnosis")
corr_data = meso_data.drop(["diagnosis"], axis=1, inplace= False)
corr_data.head()
| age | gender | duration of asbestos exposure | duration of symptoms | dyspnoea | ache on chest | weakness | habit of cigarette | cell count (WBC) | hemoglobin (HGB) | ... | glucose | pleural lactic dehydrogenise | pleural protein | pleural albumin | pleural glucose | dead or not | pleural effusion | pleural thickness on tomography | pleural level of acidity (pH) | C-reactive protein (CRP) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 47 | 1 | 20 | 24.0 | 1 | 1 | 0 | 2 | 9 | 1 | ... | 105 | 289 | 0.0 | 0.00 | 79 | 1 | 0 | 0 | 0 | 34 |
| 1 | 55 | 1 | 45 | 1.0 | 1 | 1 | 1 | 3 | 7 | 0 | ... | 96 | 7541 | 1.6 | 0.80 | 6 | 1 | 1 | 1 | 1 | 42 |
| 2 | 29 | 1 | 23 | 1.0 | 0 | 0 | 0 | 2 | 12 | 1 | ... | 93 | 480 | 0.0 | 0.00 | 90 | 1 | 0 | 0 | 0 | 43 |
| 3 | 39 | 1 | 10 | 3.0 | 0 | 1 | 1 | 0 | 14 | 1 | ... | 93 | 459 | 5.0 | 2.80 | 45 | 1 | 1 | 0 | 0 | 21 |
| 4 | 47 | 1 | 10 | 1.5 | 1 | 1 | 0 | 3 | 4 | 0 | ... | 83 | 213 | 3.6 | 1.95 | 53 | 1 | 1 | 0 | 0 | 11 |
5 rows × 26 columns
corr_mat = corr_data.corr()
plt.subplots(figsize=(15,15))
sns.heatmap(corr_mat, annot=True, cmap="YlGnBu")
<matplotlib.axes._subplots.AxesSubplot at 0x7f784b8b4690>
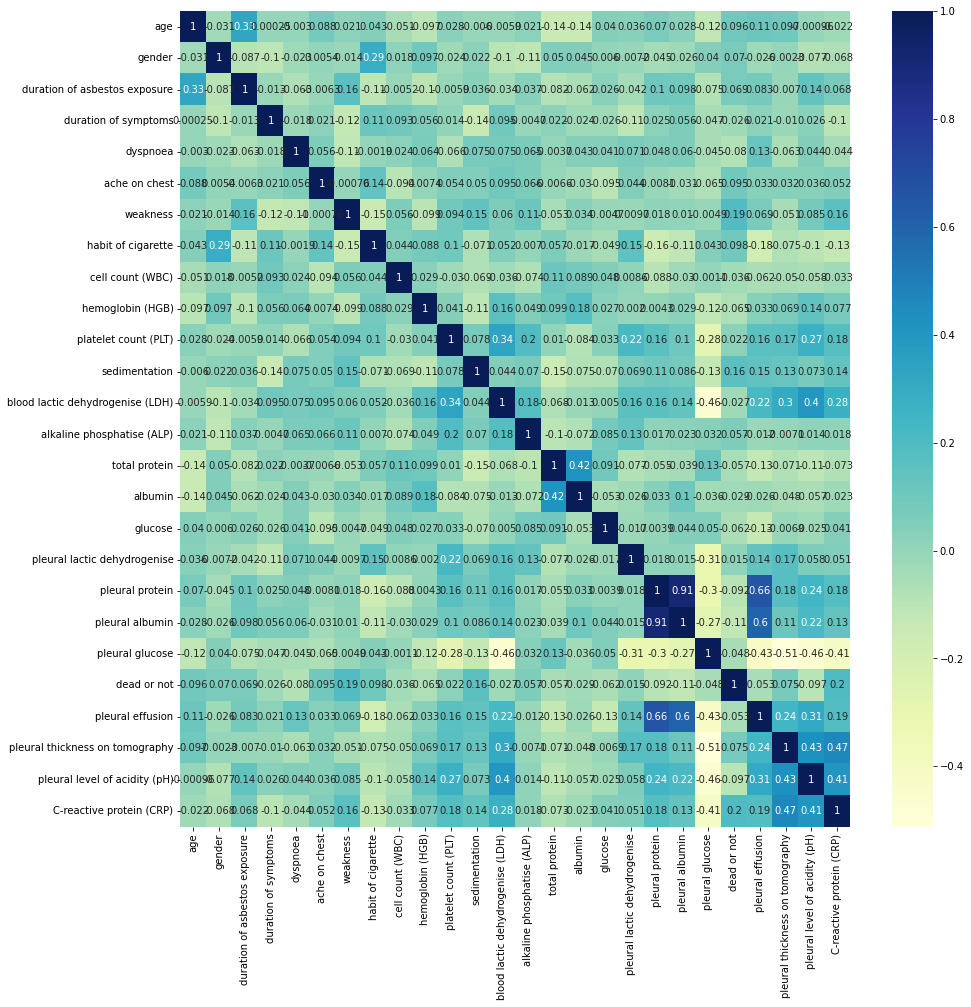
upper_tri = corr_mat.where(np.triu(np.ones(corr_mat.shape),k=1).astype(np.bool))
plt.subplots(figsize=(15,15))
sns.heatmap(upper_tri, annot=True, cmap="YlGnBu")
/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:1: DeprecationWarning:
`np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
<matplotlib.axes._subplots.AxesSubplot at 0x7f78474ace50>
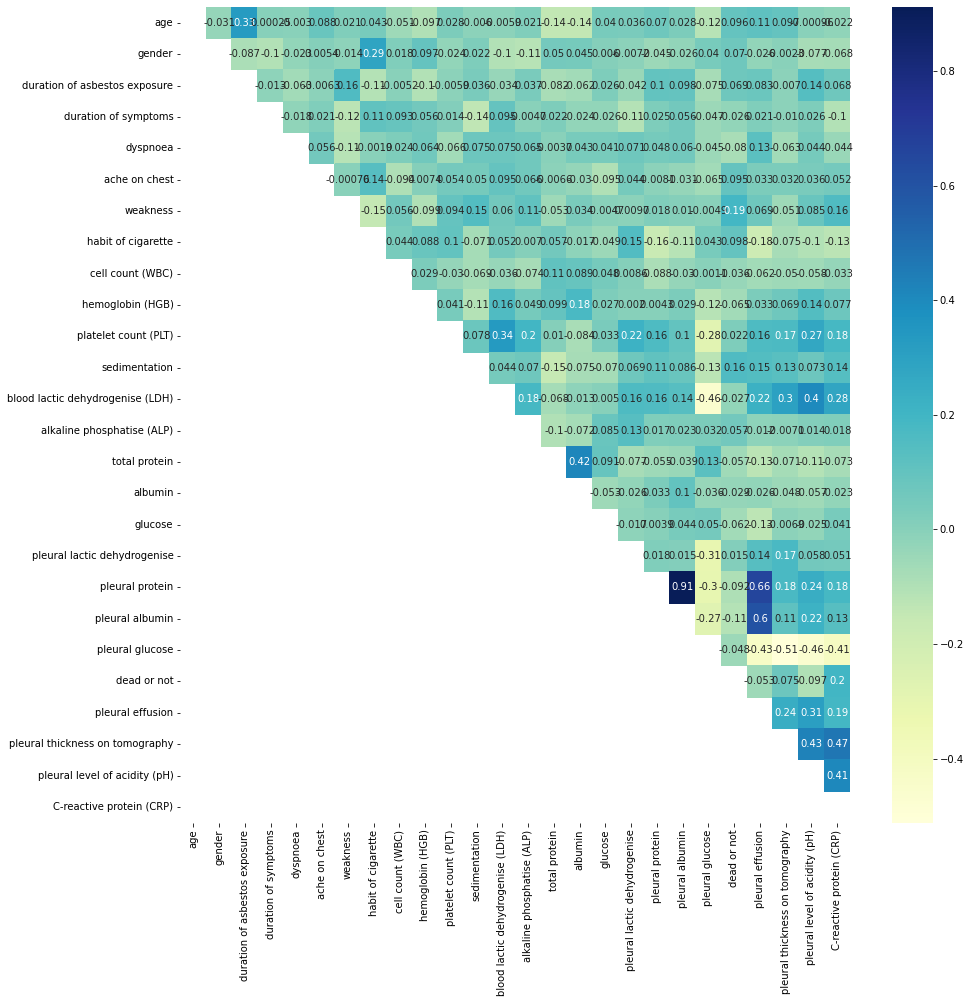
to_drop = [column for column in upper_tri.columns if any(upper_tri[column] > 0.95)]
print(); print(to_drop)
[]
Model Building
Split the data#
# Split the data
X = meso_data.iloc[:, :-1]
print(X)
print("==============================================================")
y= meso_data.iloc[:, -1]
print(y)
age gender duration of asbestos exposure duration of symptoms \
0 47 1 20 24.0
1 55 1 45 1.0
2 29 1 23 1.0
3 39 1 10 3.0
4 47 1 10 1.5
.. ... ... ... ...
319 75 1 50 9.0
320 66 1 41 9.0
321 58 1 40 8.0
322 42 1 0 2.0
323 54 1 40 3.0
dyspnoea ache on chest weakness habit of cigarette cell count (WBC) \
0 1 1 0 2 9
1 1 1 1 3 7
2 0 0 0 2 12
3 0 1 1 0 14
4 1 1 0 3 4
.. ... ... ... ... ...
319 1 1 0 2 10
320 1 1 0 2 10
321 1 1 0 2 7
322 1 0 0 3 6
323 1 1 0 3 8
hemoglobin (HGB) ... glucose pleural lactic dehydrogenise \
0 1 ... 105 289
1 0 ... 96 7541
2 1 ... 93 480
3 1 ... 93 459
4 0 ... 83 213
.. ... ... ... ...
319 0 ... 100 323
320 0 ... 100 323
321 0 ... 74 300
322 1 ... 98 3000
323 1 ... 99 2100
pleural protein pleural albumin pleural glucose dead or not \
0 0.0 0.00 79 1
1 1.6 0.80 6 1
2 0.0 0.00 90 1
3 5.0 2.80 45 1
4 3.6 1.95 53 1
.. ... ... ... ...
319 4.9 2.60 23 1
320 4.9 2.60 23 1
321 5.1 2.20 35 1
322 2.4 1.20 2 1
323 5.7 3.30 35 1
pleural effusion pleural thickness on tomography \
0 0 0
1 1 1
2 0 0
3 1 0
4 1 0
.. ... ...
319 1 1
320 1 1
321 1 0
322 1 1
323 1 1
pleural level of acidity (pH) C-reactive protein (CRP)
0 0 34
1 1 42
2 0 43
3 0 21
4 0 11
.. ... ...
319 0 76
320 0 67
321 1 68
322 0 78
323 0 45
[324 rows x 26 columns]
==============================================================
0 1
1 1
2 2
3 1
4 1
..
319 1
320 1
321 1
322 2
323 1
Name: diagnosis, Length: 324, dtype: int64
# Scale the Data
sc = StandardScaler()
X_norm = sc.fit_transform(X)
print(X_norm.shape)
(324, 26)
# Train Test valid split
# set aside 20% of train and test data for evaluation
X_train, X_test, y_train, y_test = train_test_split(X_norm, y, test_size=0.2, shuffle = True, random_state = 8)
# Use the same function above for the validation set
X_train, X_val, y_train, y_val = train_test_split(X_norm, y, test_size=0.25, random_state= 8)
print("Train data :", X_train)
print("Train data shape :", X_train.shape)
print("================================")
print("val data :", X_val)
print("================================")
print("test data :", X_test)
print("================================")
Train data : [[ 0.93359777 0.83979947 2.12336732 ... 0.82386678 0.95768458
0.83139006]
[ 0.11434676 -1.19076045 0.90345241 ... 0.82386678 -1.04418513
0.61041354]
[ 0.47845832 -1.19076045 1.39141837 ... -1.21378847 0.95768458
0.52202293]
...
[-1.1600437 -1.19076045 0.29349496 ... -1.21378847 0.95768458
1.53851492]
[ 0.29640254 0.83979947 0.23249921 ... 0.82386678 0.95768458
-0.93642211]
[-0.15873691 0.83979947 0.41548645 ... 0.82386678 0.95768458
0.74299945]]
Train data shape : (243, 26)
================================
val data : [[-0.70490425 0.83979947 -0.62144122 ... -1.21378847 -1.04418513
-1.33417984]
[ 0.11434676 0.83979947 -1.84135612 ... -1.21378847 -1.04418513
-1.24578923]
[ 0.84256988 0.83979947 0.23249921 ... 0.82386678 -1.04418513
0.1684605 ]
...
[ 0.38743043 0.83979947 -1.84135612 ... 0.82386678 -1.04418513
-1.33417984]
[-0.06770902 0.83979947 0.23249921 ... -1.21378847 -1.04418513
-0.8922268 ]
[ 0.11434676 0.83979947 0.3544907 ... 0.82386678 0.95768458
0.61041354]]
================================
test data : [[-0.70490425 0.83979947 -0.62144122 ... -1.21378847 -1.04418513
-1.33417984]
[ 0.11434676 0.83979947 -1.84135612 ... -1.21378847 -1.04418513
-1.24578923]
[ 0.84256988 0.83979947 0.23249921 ... 0.82386678 -1.04418513
0.1684605 ]
...
[ 0.75154199 -1.19076045 1.51340986 ... -1.21378847 0.95768458
1.71529614]
[-0.70490425 0.83979947 -1.84135612 ... -1.21378847 0.95768458
0.12426519]
[-1.43312737 0.83979947 -1.23139867 ... -1.21378847 -1.04418513
-1.9087188 ]]
================================
PCA#
# PCA to visualize data
from sklearn.decomposition import PCA
sklearn_pca = PCA(n_components=2)
PCs = sklearn_pca.fit_transform(X_norm)
data_transform = pd.DataFrame(PCs,columns=['PC1','PC2'])
data_transform = pd.concat([data_transform,meso_data.iloc[:,-1]],axis=1)
#plot
fig, axes = plt.subplots(figsize=(10,8))
sns.set_style("whitegrid")
sns.scatterplot(x='PC1',y='PC2',data = data_transform,hue='diagnosis',s=60, cmap='grey')
Model Building & Training#
KNN model#
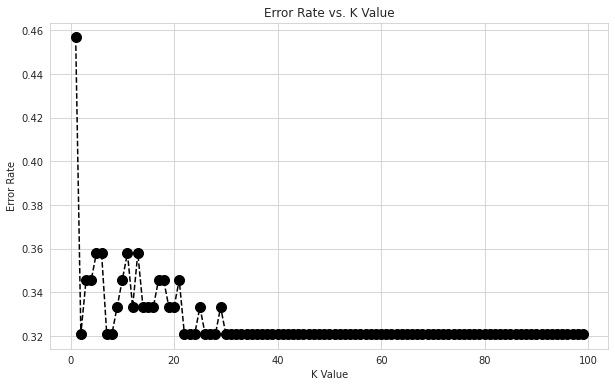
# Choose n-neigbour for KNN model
from sklearn.neighbors import KNeighborsClassifier
errors = []
for i in range(1,100):
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(X_train,y_train)
pred_i = knn.predict(X_val)
errors.append(np.mean(pred_i != y_val))
plt.figure(figsize=(10,6))
plt.plot(range(1,100),errors,color='black', linestyle='dashed',marker='o',markerfacecolor='black', markersize=10)
plt.title('Error Rate vs. K Value')
plt.xlabel('K Value')
plt.ylabel('Error Rate')
Text(0, 0.5, 'Error Rate')
Different model selection#
# Models
LogisticRegressionModel = LogisticRegression(penalty='l2',solver='sag',C=1.0,random_state=33)
lr_pred = LogisticRegressionModel.fit(X_train,y_train).predict(X_val)
KNN_classifierModel = KNeighborsClassifier(n_neighbors = 30)
KNN_pred=KNN_classifierModel.fit(X_train,y_train).predict(X_val)
param_grid = {'C': [0.1,1, 10, 100, 1000], 'gamma': [1,0.1,0.01,0.001,0.0001], 'kernel': ['rbf']}
SVM_classifier = GridSearchCV(SVC(),param_grid,refit=True,verbose=0)
#SVCModel = SVC(kernel= 'rbf', max_iter=100,C=1.0,gamma='auto')
svm_pred=SVM_classifier.fit(X_train,y_train).predict(X_val)
GaussianNBModel = GaussianNB()
gnb_pred = GaussianNBModel.fit(X_train,y_train).predict(X_val)
DecisionTreeClassifierModel = DecisionTreeClassifier(criterion='entropy',max_depth=3,random_state=33)
dt_pred = DecisionTreeClassifierModel.fit(X_train,y_train).predict(X_val)
param_grid = {'n_estimators': [10, 100,150,200,250,300,350,400]}
RandomForestClassifierModel = GridSearchCV(RandomForestClassifier(),param_grid,refit=True, verbose=0)
#RandomForestClassifierModel = RandomForestClassifier(criterion = 'gini',n_estimators=100,max_depth=2,random_state=33)
rf_pred = RandomForestClassifierModel.fit(X_train,y_train).predict(X_val)
SGDClassifierModel = SGDClassifier(penalty='l2',learning_rate='optimal',random_state=33)
SGD_pred = SGDClassifierModel.fit(X_train,y_train).predict(X_val)
GBCModel = GradientBoostingClassifier(n_estimators=100,max_depth=3,random_state=33)
GBC_pred = GBCModel.fit(X_train,y_train).predict(X_val)
print(X_train.shape[1])
26
Artificial Neural Network#
#ANN
from tensorflow.keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, TensorBoard
from datetime import datetime
filepath="/content/drive/MyDrive/Mesothelioma/weights-improvement-{epoch:02d}-{val_loss:.12f}.hdf5"
log_dir = "/content/drive/MyDrive/Mesothelioma/"+datetime.now().strftime("%Y%m%d-%H%M%S")
checkpoint = ModelCheckpoint(filepath, monitor='val_accuracy', verbose=1, save_best_only=True, mode='max')
reduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.2, patience=5, min_lr=0.0001)
tensorboard = TensorBoard(log_dir="./logs", histogram_freq=1, write_graph=True)
# determine the number of input features
n_features = X_train.shape[1]
print("The Number of Features (columns) : ", n_features )
# define model
ANN_model = Sequential()
ANN_model.add(Dense(26, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,)))
ANN_model.add(Dropout(0.5))
ANN_model.add(Dense(26, activation='relu', kernel_initializer='he_normal'))
ANN_model.add(Dropout(0.5))
ANN_model.add(Dense(1, activation='sigmoid'))
ANN_model.summary()
# compile the model
ANN_model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# Early stopping
early_stop = EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=5 )
# call backs
callbacks= [checkpoint, tensorboard, reduce_lr]
# Fit
history=ANN_model.fit(x=X_train, y=y_train, epochs=400, batch_size= 128, validation_split= 0.2, verbose=0, callbacks= callbacks )
# evaluate the model
loss, acc_ANN = ANN_model.evaluate(X_val, y_val, verbose=0)
print('Test Accuracy: %.3f' % acc_ANN)
ANN_pred = ANN_model.predict(X_val)
print(ANN_pred)
ANN_model_performance = pd.DataFrame(ANN_model.history.history)
#ax.set_xlabel('Epoch')
print(ANN_model_performance.head())
ANN_model_performance.plot()
Prediction#
print(lr_pred)
[2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 2 2 1 1 2
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 2 2 2 1 1 1 1 2 1 1 1 1 1 2 1 2
2 1 1 1 1 1 1]
history.history.keys()
dict_keys(['loss', 'accuracy', 'val_loss', 'val_accuracy', 'lr'])
ANN_acc= history.history["accuracy"]
print(ANN_acc)
[0.5, 0.4793814420700073, 0.530927836894989, 0.5206185579299927, 0.5257731676101685, 0.5824742317199707, 0.5876288414001465, 0.5979381203651428, 0.6134020686149597, 0.5721649527549744, 0.5979381203651428, 0.5773195624351501, 0.5979381203651428, 0.592783510684967, 0.6185566782951355, 0.6340206265449524, 0.6185566782951355, 0.623711347579956, 0.6443299055099487, 0.6443299055099487, 0.6546391844749451, 0.6494845151901245, 0.6546391844749451, 0.6804123520851135, 0.6546391844749451, 0.6649484634399414, 0.6752577424049377, 0.6649484634399414, 0.6752577424049377, 0.7010309100151062, 0.6907216310501099, 0.6855670213699341, 0.6701030731201172, 0.6907216310501099, 0.6958763003349304, 0.6752577424049377, 0.6855670213699341, 0.6958763003349304, 0.6958763003349304, 0.7061855792999268, 0.7010309100151062, 0.7010309100151062, 0.7061855792999268, 0.7061855792999268, 0.7010309100151062, 0.7010309100151062, 0.6907216310501099, 0.7010309100151062, 0.7010309100151062, 0.7061855792999268, 0.7113401889801025, 0.6958763003349304, 0.7061855792999268, 0.7061855792999268, 0.7010309100151062, 0.7113401889801025, 0.7113401889801025, 0.7010309100151062, 0.7113401889801025, 0.7061855792999268, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7061855792999268, 0.7113401889801025, 0.7061855792999268, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7061855792999268, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025, 0.7113401889801025]
models=['Logistic Regression','KNN', 'SVM','GaussianNB','DecisionTree Classifier','RandomForest Classifier','SGD Classifier','GBCModel']
preds=[lr_pred,KNN_pred, svm_pred,gnb_pred,dt_pred,rf_pred,SGD_pred,GBC_pred,y ]
print(preds)
# models2=['Logistic Regression','KNN', 'SVM','GaussianNB','DecisionTree Classifier','RandomForest Classifier','SGD Classifier','GBCModel', "ANN"]
# preds=[lr_pred,KNN_pred, svm_pred,gnb_pred,dt_pred,rf_pred,SGD_pred,GBC_pred,y ]
# print(preds)
print(lr_pred)
[2 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 2 2 1 1 2
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 2 2 2 1 1 1 1 2 1 1 1 1 1 2 1 2
2 1 1 1 1 1 1]
ANN_pred
from itertools import chain
x = ANN_pred
y = list(chain(*x))
print(y)
[1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0]
acc=[]
for i in preds:
accscore=accuracy_score(i,y_val).round(2)
acc.append(accscore)
print(acc)
join = zip(models, acc)
print(join)
result = pd.DataFrame(join, columns=['model', 'accuracy']).sort_values(['accuracy'], ascending=False)
print(result)
Plotting Results#
sns.barplot(x = 'model', y= 'accuracy', data= result), plt.xticks(rotation =-90)
